Today, Adrien Boussebayle explains the focus of his PhD to us: aptamers. And admits some lab secrets…
What a strange word… Aptamer comes from the Latin “aptāre “ (to put into position, fit together, join), and from the Greek “meros” (part). By definition, an aptamer is a single-strand DNA or RNA which can bind a specific target with high specificity (uniqueness of binding) and high affinity (strength of binding). These targets can be small, like a Zn2+ ion or as big as protein and even entire cells.
Aptamer are selected by a process called SELEX (Systematic Evolution of Ligand by Exponential Enrichment) by combining your desired binding target in contact with a large number of potential aptamers. The researcher constructs a large RNA or DNA aptamer pool, also known as a SELEX library, composed of several tens (usually 30 to 80) of random nucleotides. Overall, this selection library will contain 1013 to 1016 unique pieces of RNA or DNA.

This pool of potential aptamers is incubated with the binding target, which is bound to a solid matrix. The matrix is used to filter out low-affinity or non-specific binders. The remaining sequences will have some binding properties. The selected aptamers are amplified (i.e. multiple copies are made) and used in the next SELEX round. Each round will produce a smaller aptamer pool with improves binding properties.
Aptamers can assume a variety of shapes like stems, loops or helices because of their nucleic acid backbone. This explains the versatility of aptamers to bind to very diverse targets, as you can see in this image:
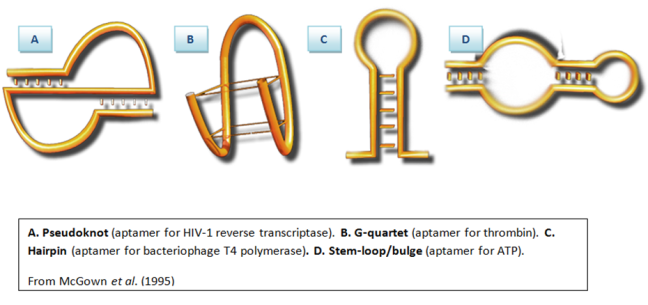
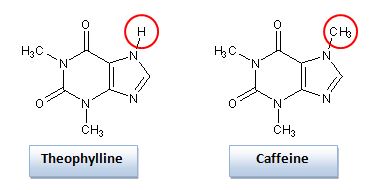
Traditionally, antibodies have been used to target specific molecules. However, aptamers provide a number advantages over traditional antibodies. In many cases, aptamers offer much higher binding affinity. The dissociation constant (KD) is a way to quantify binding ability and for some aptamers, this can span from the µM to the pM range. Likewise, aptamers are very good at target discrimination. For example, the aptamer for theophylline has 10,000-fold less binding affinity for caffeine, even though caffeine and theophylline differ only by a methyl group! You can see the molecular similarity in the figure below.
Not only that, but antibodies are larger and more fragile compounds that can cause undesired immunogenic responses. As you can see in the image below, aptamers are 5 times smaller, very stable**, and unlikely to interfere with the immune response.
** Once, I forgot a G4 DNA aptamer against integrase for one week on my bench, during summer in the south-west of France (35°C outside). It did not seem to “hurt” it. Thankfully for my life, no one noticed it. That’s why working with nucleic acid is cool – I have less chance to annihilate my sample.
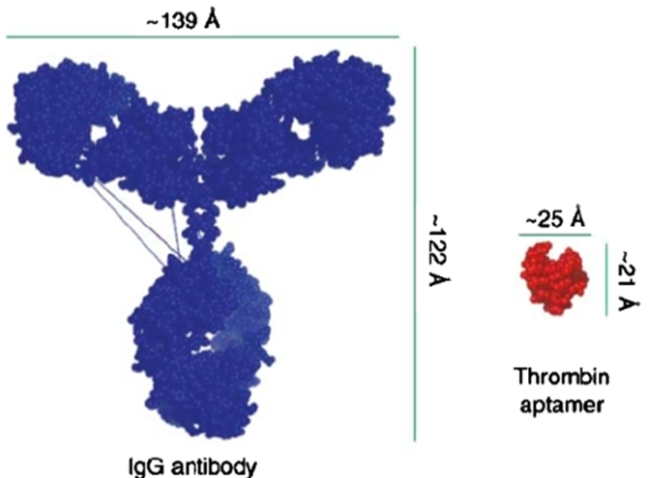
Image from “Aptamer therapeutics advance” by Jennifer F Lee, Gwendolyn M Stovall and Andrew D Ellington. Current Opinion in Chemical Biology, 2006.
All these advantages means aptamers make great biosensors, which are constructs that sense what is going on the cell and provide the researcher with information. Indeed, aptamer size, stability, high affinity and specificity are exactly the properties you want in a biosensor! This bring us to the purpose of MetaRNA: Developing RNA-based sensors to monitor metabolic fluxes.
Next week, Adrien will tell us about one of the most well-known aptamer with the intriguing name of Spinach!

One thought on “What are aptamers?”